About our Research
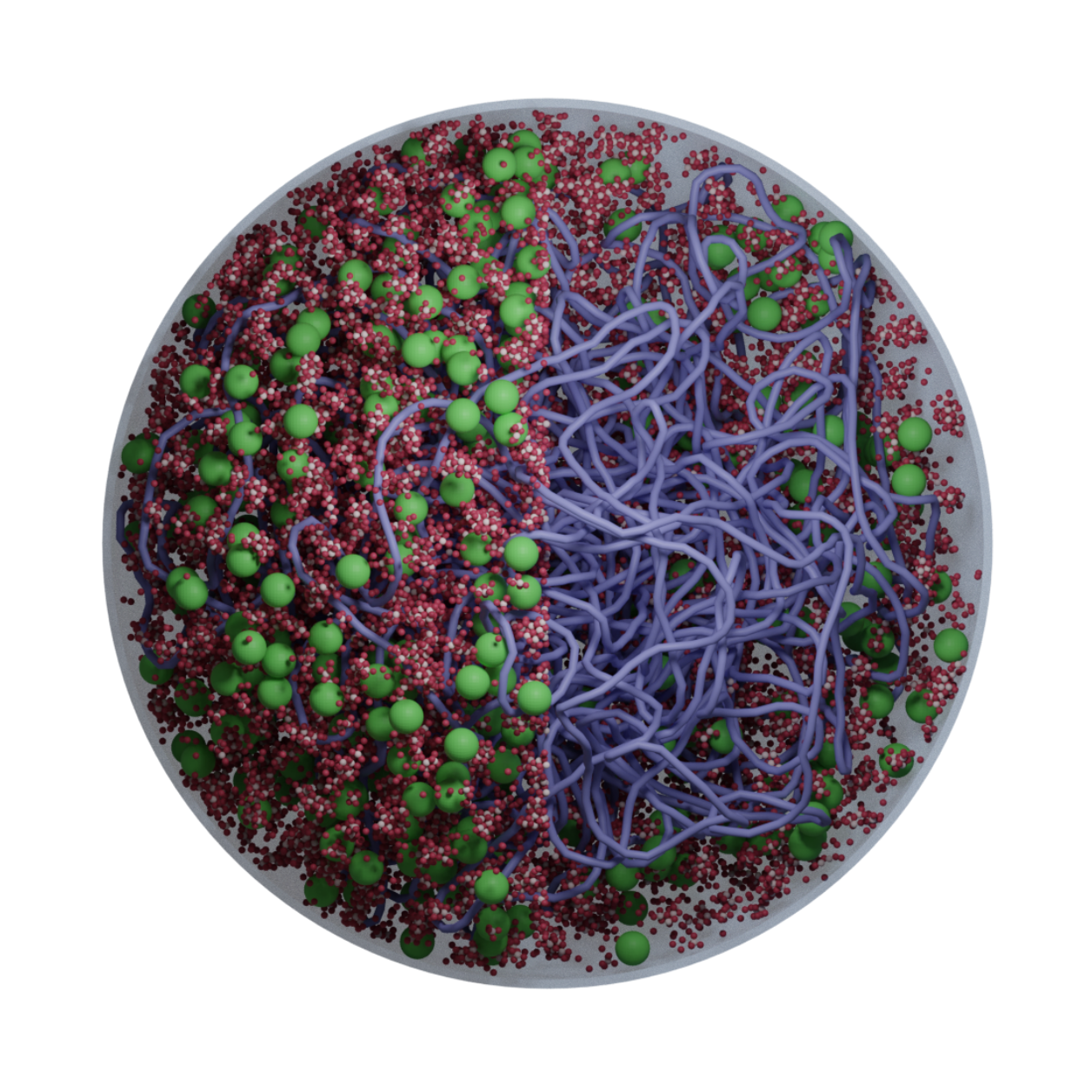
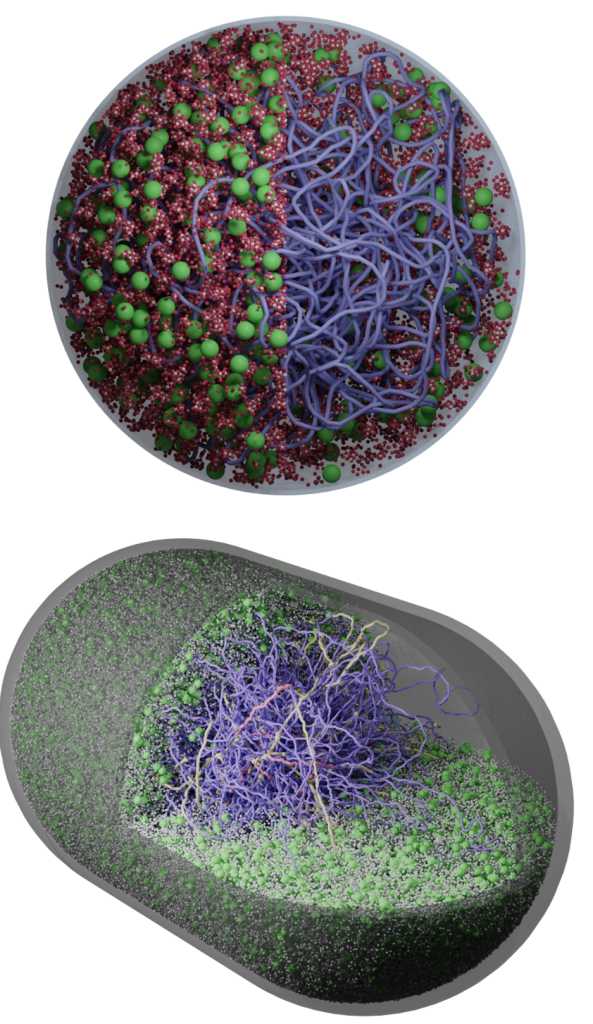
We develop AI-guided, physics-based whole-cell models of bacteria and synthetic cells using tools from physics, cell biology, and scientific computing. These models preserve near-atomistic resolution for millions of biomolecules as they dynamically interact, letting us watch transcription, translation, ribogenesis, and more unfold inside a functioning in silico cell.
We use these models to uncover how cell-scale physics, spatial architecture, and physicochemical processes control cell function and how matter becomes life, connecting physiotype to phenotype—cytoplasmic organization and nucleoid restructuring to gene expression and adaptation. A central focus is ribosome biogenesis and engineering in natural and synthetic cells.
In parallel, we study colloidal gels, phase separation, and soft-matter rheology, from entropy-driven crystallization in model colloids to the stability and flow of protein suspensions and gels. These systems provide both a testing ground for our methods and a bridge between idealized colloids and the crowded cytoplasm inside living cells.
Research Topics
Whole-cell models
Engineering biological cells as programmable factories underpins future advances in medicine, biosecurity, and designed biomaterials. To get there, we need models that capture how real cells work in space and time, not just lists of reactions.
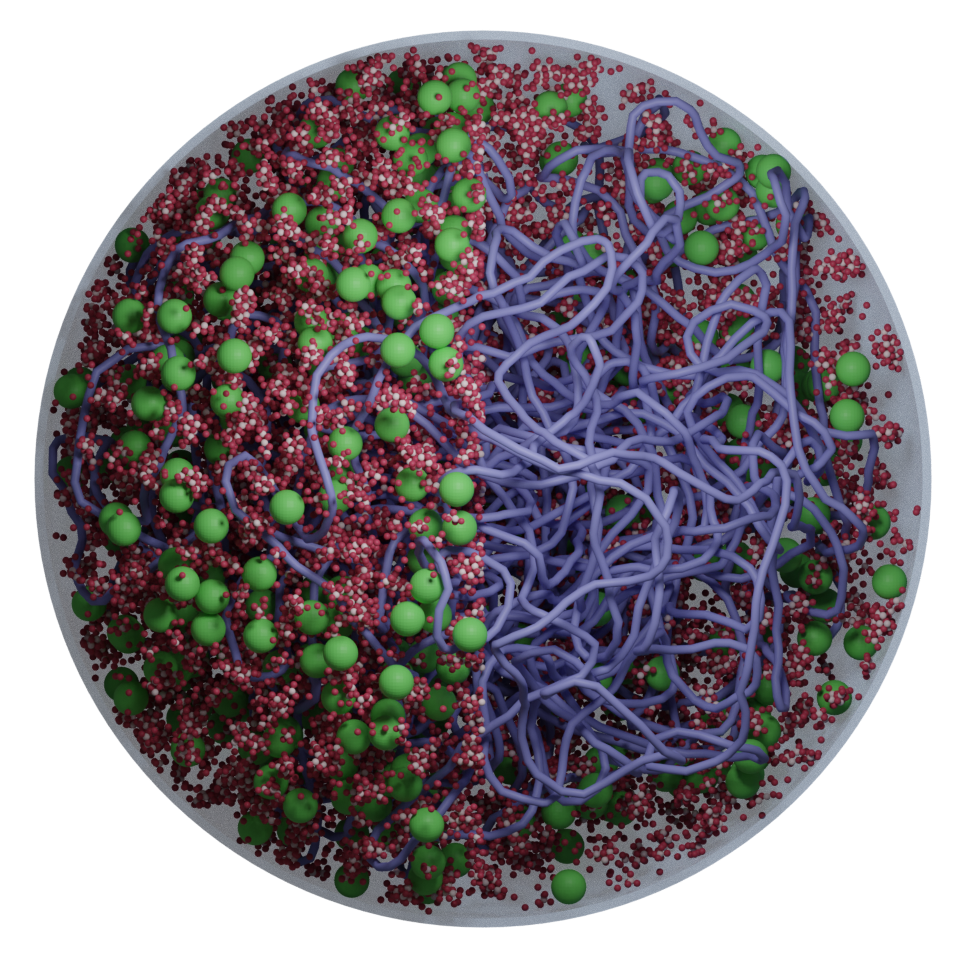
Classic whole-cell models track only reaction kinetics and ignore spatial organization; brute-force all-atom simulations are so expensive that even a few reactions are impractical. We bridge these extremes with an AI-guided physics pipeline: near-atomistic GROMACS simulations are used to learn how biomolecules interact, and those data train coarse-grained representations whose particles retain near-atomistic interaction behavior. This lets our colloidal- scale Brownian dynamics simulate millions of biomolecules with fast, on-the-fly reaction computation inside a functioning in silico cell.
Ribosome Biosynthesis
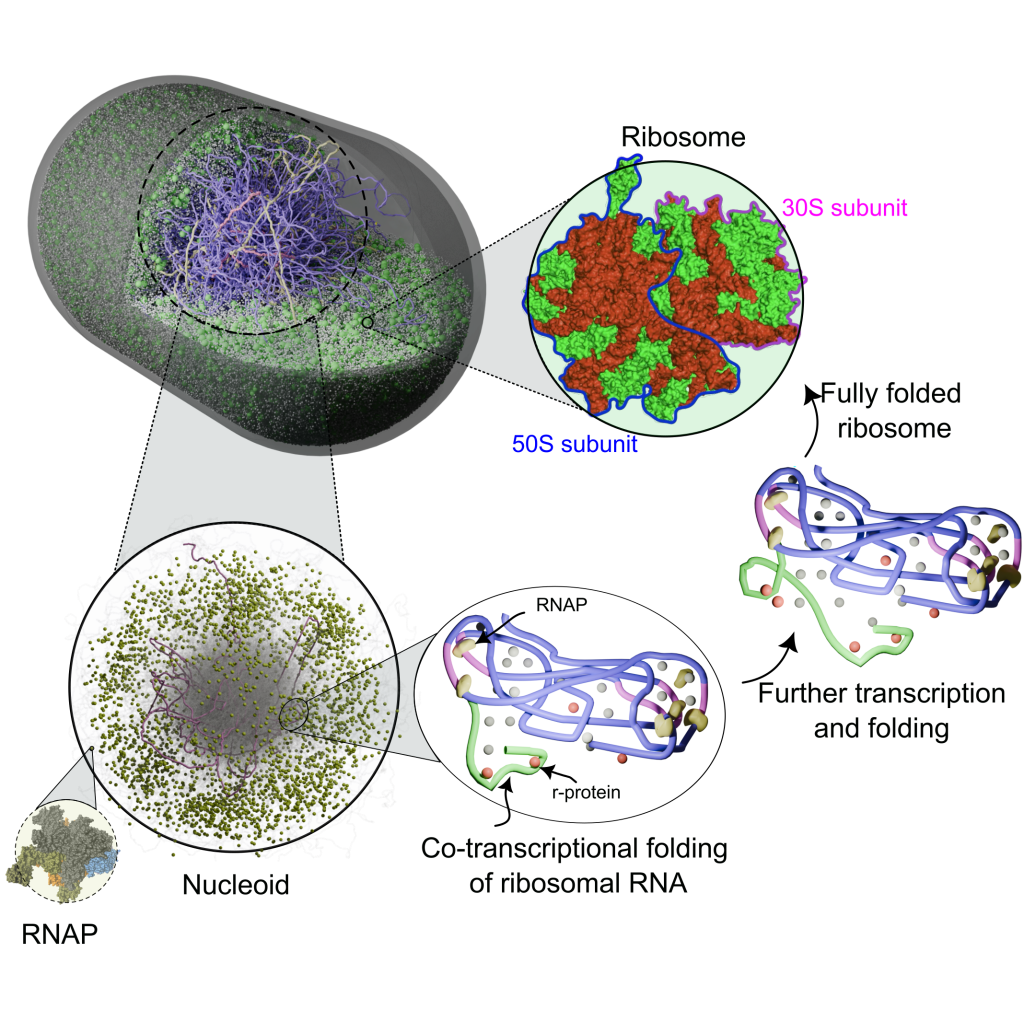
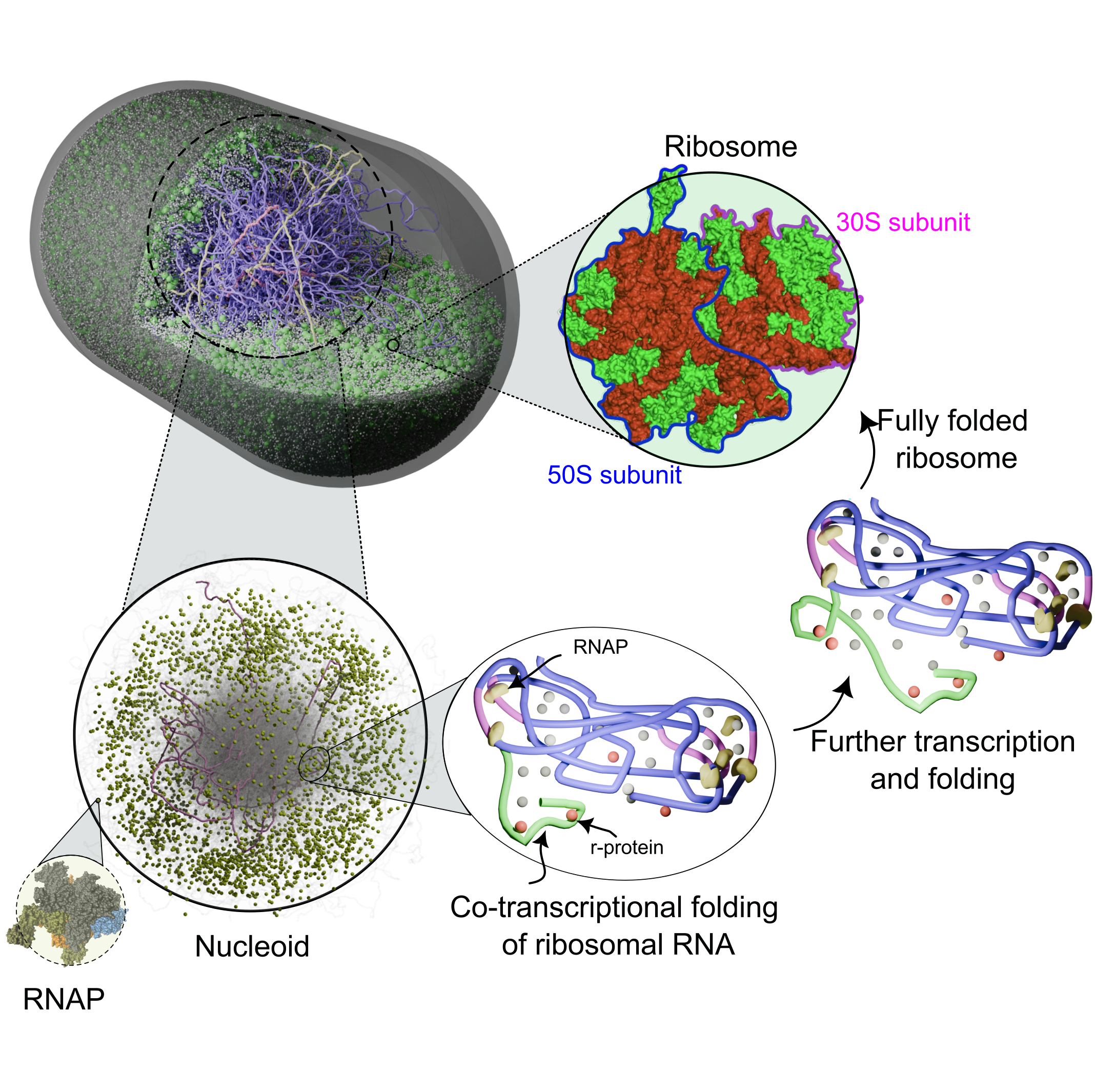
We study how cells build ribosomes from first principles, using physics-based, AI-guided whole-cell models of bacteria and synthetic cells. Our simulations explicitly track rRNA transcription, nucleoid restructuring, ribosomal protein synthesis, and stepwise assembly of ribosomal subunits in the crowded cytoplasm. By coupling genome organization, transcriptional dynamics, and colloidal-scale interactions among ribosomal components, we probe how cells accelerate ribosome production under fast growth and how minimal cells sustain autonomous ribogenesis. A key goal is to extract the physical design rules for ribosome biosynthesis—linking genome architecture, spatial organization, and assembly pathways—and use them to guide engineering of synthetic cells and, ultimately, custom ribosomes.
Colloidal Phyiscs & Rheology
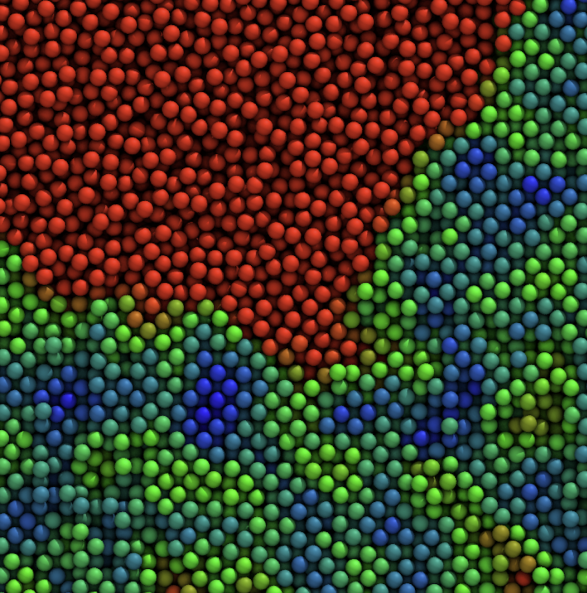
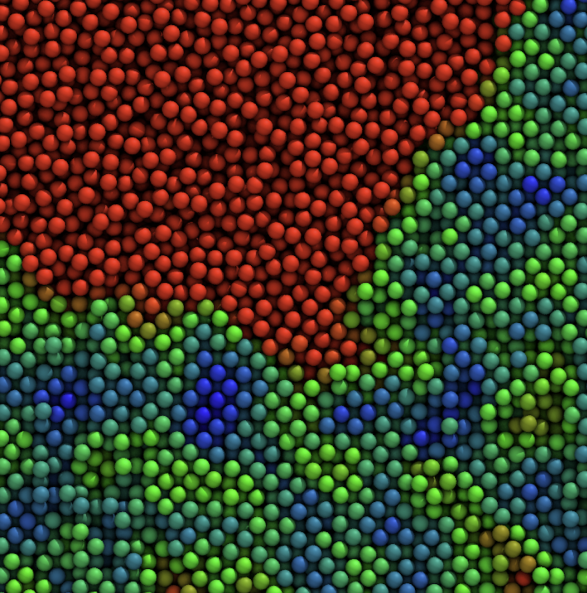
We study how colloidal particles—from simple spheres to complex proteins—organize, jam, and flow, and how this controls the mechanics of gels, glasses, and protein formulations. On the fundamental side, we use large-scale simulations to probe entropy-driven phase separation, showing how small changes in particle “hardness” unlock thermodynamically guaranteed but kinetically elusive states.
On the applied side, we build multiscale models of globular and therapeutic proteins with experimental partners, bridging sequence-level detail into efficient colloidal models that predict suspension stability, gelation, and viscoelastic response. We use these tools to study prion-like aggregation implicated in epigenetic inheritance.
We also tackle colloidal phase (mis)behavior: sudden gel collapse, colloidal glasses, and how aging rewrites structure–property relationships in soft materials.